Data Matrices Construction¶
Extracted data are organized in order to use them as input for the data analysis. The single regulation system of each target gene is analyzed, by identifying the role of its possible regulatory features. They are quantified for their impact on the target expression, whatever the feature is, i.e. the gene methylation, the expression of a gene in the same gene set or in another gene set and the expression of genes encoding for transcription factors.
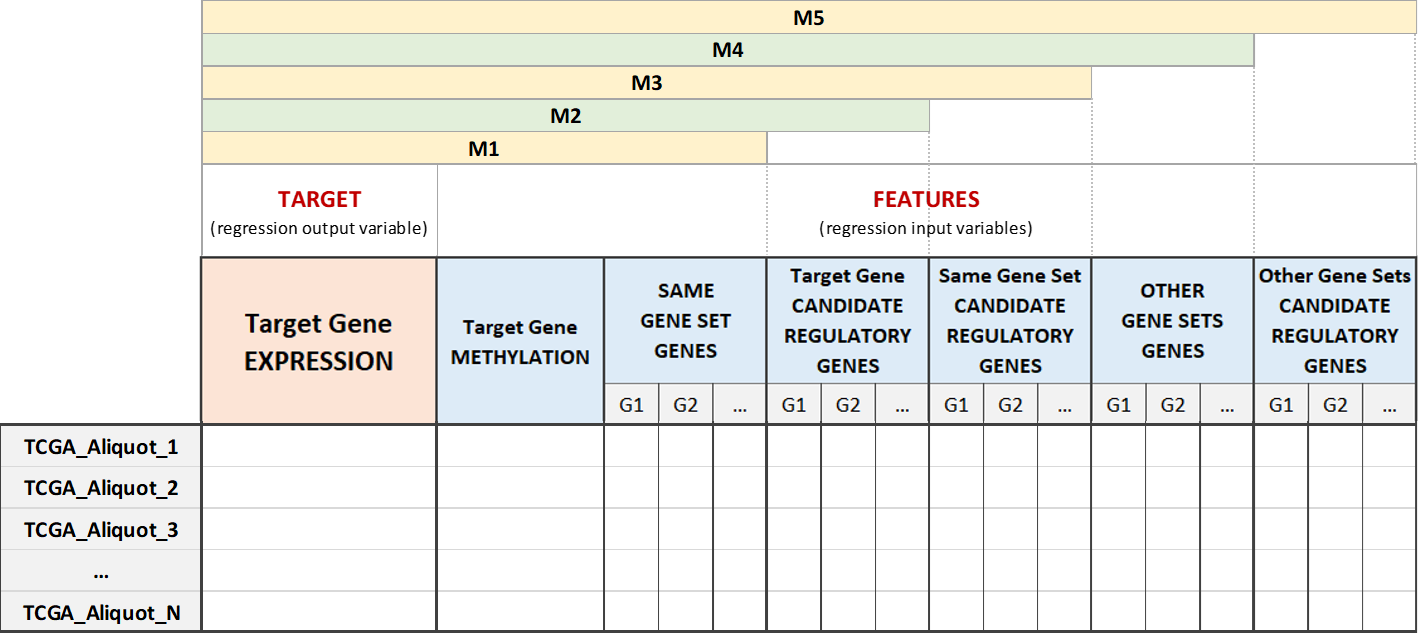
An additive approach is adopted, building five different data matrices for each gene of interest. In this way it is possible to keep track of the components of each gene regulation system step by step, according to the different types of biological features. More specifically, the matrices are built starting from the gene expression and methylation value of the target gene and gradually incrementing the number of columns/features, as follows:
- Matrix M1 contains the expression of the target gene, its methylation and the expression of the genes belonging to the same gene set as the target gene;
- Matrix M2 adds the expression of all the candidate regulatory genes of the target gene to M1, avoiding repetitions;
- Matrix M3 adds the expression of the candidate regulatory genes of all the genes in the target gene set to M2, avoiding repetitions;
- Matrix M4 adds the expression of the genes belonging to other gene sets with respect to the considered target gene to matrix M3, avoiding repetitions;
- Matrix M5 adds the expression of the candidate regulatory genes of all the genes belonging to other gene sets to matrix M4, avoiding repetitions.
Here, it is the set of functions used to build the five data matrices:
create_m1()
The CREATE_M1 operation builds the first data matrix for each target gene of interest, collecting the target gene expression and methylation values, along with the expression values of all the genes in the same gene set. One data matrix for each target gene is created and exported locally in as many Excel files as the considered target genes; while the whole set of M1 matrices is returned as a Python dictionary (dict_model_v1.p), where each target gene (set as key) is associated with a Pandas dataframe containing the data for its M1 matrix (set as value).
Return: a Python dictionary
INPUT FILES: Genes_of_Interest.xlsx from ./, Methylation_Values.xlsx from ./3_TCGA_Data/Methylation/, Gene_Expression-InterestGenes.xlsx from ./3_TCGA_Data/Gene_Expression/
OUTPUT_FILES: dict_model_v1.p, Gene_ID_[SYMBOL]_(gene_set)-Model_v1.xlsx in ./4_Data_Matrix_Construction/Model1/ (click on the Excel file to download a sample matrix for a sample)
Example:
import genereg as gr m1_dict = gr.DataMatrices.create_m1()
create_m2()
The CREATE_M2 operation builds the second data matrix for each target gene of interest, adding to the first matrix data about the expression of candidate regulatory genes of each target gene of interest. One data matrix for each target gene is created and exported locally in as many Excel files as the considered target genes; while the whole set of M2 matrices is returned as a Python dictionary (dict_model_v2.p), where each target gene (set as key) is associated with a Pandas dataframe containing the data for its M2 matrix (set as value).
Return: a Python dictionary
INPUT FILES: Genes_of_Interest.xlsx from ./, dict_RegulGenes.p from ./2_Regulatory_Genes/, Gene_Expression-RegulatoryGenes.xlsx from ./3_TCGA_Data/Gene_Expression/, dict_model_v1.p from ./4_Data_Matrix_Construction/Model1/
OUTPUT_FILES: dict_model_v2.p, Gene_ID_[SYMBOL]_(gene_set)-Model_v2.xlsx in ./4_Data_Matrix_Construction/Model2/ (click on the Excel file to download a sample matrix for a sample)
Example:
import genereg as gr m2_dict = gr.DataMatrices.create_m2()
create_m3()
The CREATE_M3 operation builds the third data matrix for each target gene of interest, adding to the second matrix data about the expression of candidate regulatory genes of genes of interest belonging to the same gene set of the target gene. One data matrix for each target gene is created and exported locally in as many Excel files as the considered target genes; while the whole set of M3 matrices is returned as a Python dictionary (dict_model_v3.p), where each target gene (set as key) is associated with a Pandas dataframe containing the data for its M3 matrix (set as value).
Return: a Python dictionary
INPUT FILES: Genes_of_Interest.xlsx from ./, dict_RegulGenes.p from ./2_Regulatory_Genes/, Gene_Expression-RegulatoryGenes.xlsx from ./3_TCGA_Data/Gene_Expression/, dict_model_v2.p from ./4_Data_Matrix_Construction/Model2/
OUTPUT_FILES: dict_model_v3.p, Gene_ID_[SYMBOL]_(gene_set)-Model_v3.xlsx in ./4_Data_Matrix_Construction/Model3/ (click on the Excel file to download a sample matrix for a sample)
Example:
import genereg as gr m3_dict = gr.DataMatrices.create_m3()
create_m4()
The CREATE_M4 operation builds the fourth data matrix for each target gene of interest, adding to the third matrix data about the expression of genes of interest belonging to other gene sets with respect to the target gene. One data matrix for each target gene is created and exported locally in as many Excel files as the considered target genes; while the whole set of M4 matrices is returned as a Python dictionary (dict_model_v4.p), where each target gene (set as key) is associated with a Pandas dataframe containing the data for its M4 matrix (set as value).
Return: a Python dictionary
INPUT FILES: Genes_of_Interest.xlsx from ./, Gene_Expression-InterestGenes.xlsx from ./3_TCGA_Data/Gene_Expression/, dict_model_v3.p from ./4_Data_Matrix_Construction/Model3/
OUTPUT_FILES: dict_model_v4.p, Gene_ID_[SYMBOL]_(gene_set)-Model_v4.xlsx in ./4_Data_Matrix_Construction/Model4/ (click on the Excel file to download a sample matrix for a sample)
Example:
import genereg as gr m4_dict = gr.DataMatrices.create_m4()
create_m5()
The CREATE_M5 operation builds the fifth data matrix for each target gene of interest, adding to the fourth matrix data about the expression of candidate regulatory genes of genes of interest belonging to other gene sets with respect to the target gene. One data matrix for each target gene is created and exported locally in as many Excel files as the considered target genes; while the whole set of M5 matrices is returned as a Python dictionary (dict_model_v5.p), where each target gene (set as key) is associated with a Pandas dataframe containing the data for its M5 matrix (set as value).
Return: a Python dictionary
INPUT FILES: Genes_of_Interest.xlsx from ./, dict_RegulGenes.p from ./2_Regulatory_Genes/, Gene_Expression-RegulatoryGenes.xlsx from ./3_TCGA_Data/Gene_Expression/, dict_model_v4.p from ./4_Data_Matrix_Construction/Model4/
OUTPUT_FILES: dict_model_v5.p, Gene_ID_[SYMBOL]_(gene_set)-Model_v5.xlsx in ./4_Data_Matrix_Construction/Model5/ (click on the Excel file to download a sample matrix for a sample)
Example:
import genereg as gr m5_dict = gr.DataMatrices.create_m5()